Just published yesterday in the early edition of the Proceedings of the National Academy of Sciences is a tour de force of thinking big, working together, and demonstrating that even science that is messy and incomplete can be incredibly useful and worthy of publication.
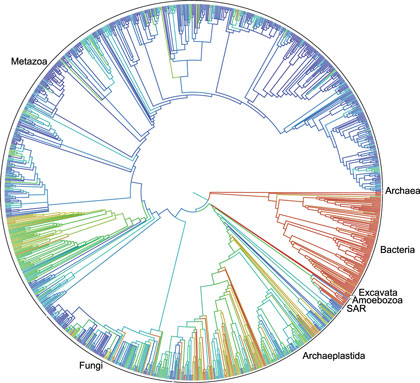
In an article with the dense, yet understated, title “Synthesis of phylogeny and taxonomy into a comprehensive tree of life”, twenty-two authors from thirteen institutions (supported by a grant from the National Science Foundation) present their initial effort at building a single tree to model how all living things on Earth are related. Their tree is available to all, on-line, in an open source format, so that everyone can explore it freely and researchers can add their own data.
The project made widespread headlines this week, from Science News to CNET to The Christian Science Monitor. But why? Why is a crazy complicated looking diagram such a big deal? Well, for one thing, trees are important. Understanding how one organism is related to all others is important in biology for so many reasons. Whether you’re looking for a potential drought-resistant crop, a more potent version of an antibiotic, the best model organism to study a particular disease, or the most promising algal strain to cultivate for biofuels, it’s helpful to be able to look for clues in closely related species. Trees are so useful that they’ve become a standard part of biological investigations—countless phylogenies (trees generated by organizing species according to the traits they share) have been generated for all manner of organisms.
Here’s the thing, though. As Stephanie Keep so ably explained in her four part “Do You Talk Tree?” series (here, and here, and here, and here), phylogenetic trees are models. They are generated by computer programs with many embedded choices, and changing the weighting of various parameters can result in trees with slightly, or dramatically, different shapes. Furthermore, trees that are generated using different genes or sets of genes may give different answers as to which species are most closely related. Oh, and remember that not all trees are based on gene sequences; trees that compare morphological differences are also important (indeed, for fossils, they’re pretty much the only game in town). And finally, the same tree can be drawn in many different ways, depending on what the researcher is trying to emphasize.
So how do you go about making one, massive, tree of life? One approach would have been essentially to start over—use information directly from sequence databases to generate a comprehensive tree of life from scratch, as it were. This would, however, have ignored the literally thousands of careful and detailed phylogenies that have been generated over the decades. So instead, the researchers chose another tack and developed a program that pieces those phylogenies together into a single tree. The task of combining thousands of phylogenetic trees into one big, comprehensive tree of life, turned out to be enormously challenging, so kudos are due the authors for pulling it off (and to the peer reviewers at the National Science Foundation who recommended funding something so ambitious).
The authors are the first to admit that the version published yesterday is very much a rough draft; out of approximately 7,500 phylogenies published between 2010 and 2012, they incorporated only 484. Why so few? Well, there were problems with inconsistent species names and abbreviations, trees that didn’t agree with each other, and—the biggest problem—trees that had been published as images without the underlying data. In all, the underlying data that was used to generate the trees was available in digital form for only about one study in six. In other words: the data was messy and there were technical difficulties.
We can tell that the project isn’t finished because so few species are represented. Yes, I said few—2.3 million might sound like a lot, but it’s really a drop in the bucket. A 2011 study that estimated the total number of species on Earth at approximately 8.7 million came under considerable criticism for massive underestimates of some kinds of organisms. For example, it estimated total bacterial diversity at 10,000 species, which is about the number of bacterial species found in the average teaspoon of soil. Other groups— like single-celled eukaryotes and nematodes—have received considerably less attention than the plants and animals we can see with our own eyes and it’s likely we’ve discovered only a tiny fraction of species from these groups. So the total number of species could easily end up in the many tens, or even hundreds, of millions.
What’s great about this project, though, is that as those species are identified, and phylogenies of various groups of microbes, insects, algae, worms, diatoms and others are generated, it will be possible to incorporate those new phylogenies directly into this comprehensive, open source tree. So as we learn more, the tree will get more complete, and more useful. This work has the potential to provide a framework upon which a vast amount of biological information can be organized—information that is currently fragmented and scattered across thousands of separate studies and unavailable in searchable form. Efforts like this, that synthesize information and enable researchers to see a larger picture than is available in just their own particular corner of biology, have the potential to make biologists more efficient, and, indeed, to boost the value of all biological research.
Sure, there will be disputes, there will be messiness, occasionally there may be dramatic re-evaluations of who’s related to whom, but the important thing is that there will be a common sandbox where everyone can play. In the long run, all of the important problems that biological research addresses—in health, agriculture, energy, and environmental protection—will get just a little bit easier to solve.

