 “How extremely stupid not to have thought of that!” was Thomas Henry Huxley’s reflection on reading Darwin’s Origin of Species. What might elicit such a reaction from a contemporary biologist? Today the question is answered by David W. Deamer. Research Professor in the Chemistry and Biochemistry Department at the University of California, Santa Cruz, Deamer is the author of First Life: Discovering the Connections between Stars, Cells, and How Life Began (2012).
“How extremely stupid not to have thought of that!” was Thomas Henry Huxley’s reflection on reading Darwin’s Origin of Species. What might elicit such a reaction from a contemporary biologist? Today the question is answered by David W. Deamer. Research Professor in the Chemistry and Biochemistry Department at the University of California, Santa Cruz, Deamer is the author of First Life: Discovering the Connections between Stars, Cells, and How Life Began (2012).
“Why didn’t I think of that?” I asked myself when I read a paper by David Bartel and Jack Szostak published in Science in 1993.
Their experiment precisely refuted the idea that it is impossible for genetic information to emerge from randomness. Here’s how it was done. First, RNA molecules composed of ~300 nucleotides were synthesized by a process that assured their base sequences were random. The RNA was analyzed by gel electrophoresis, a common procedure in molecular biology. A gel is produced when a polymeric material such as agarose or polyacrylamide is mixed with water and allowed to set. If you have ever made Jell-O, that’s exactly what happens. For electrophoresis, the gel is a thin layer between two glass plates and the nucleic acid samples are added to ten or so small wells at the top of the gel. A voltage of several hundred volts is applied, and this causes the negatively charged RNA molecules to move through the gel from top to bottom. Small molecules move fastest, while longer molecules move more slowly. If the RNA molecules have an exact length, they all travel at the same rate and produce a tight band on the gel. The random sequence RNA molecules in Bartel’s experiment were 300 nucleotides long, so they produced the band seen at the top of the gel in the figure below.
The next step was to choose something to select for, which could be almost any function one might imagine RNA could perform. Bartel and Szostak chose a function called ligation, which is the catalyzed synthesis of a chemical bond between the ends of two long molecules. The idea was to bind the RNA to beads and then expose the beads to a short substrate RNA that had the potential to be ligated. The mixture of random RNA molecules was released from the beads, and those with ligase activity would have the short strand of RNA attached. These were separated and amplified by standard methods, then the second generation of RNA molecules would be put through the same iterative process.
This laboratory experiment seems far from what we usually call evolution, but in fact it can be compared to selecting the smallest puppies out of a litter, letting them grow up and breed and then again selecting the smallest puppies out of the next litter. Or in the natural world, it is like peahens being attracted to peacocks with the most colorful tail feathers. The dog breeder ends up with chihuahuas, and the tails of peacocks become ever more colorful over many generations, but what happened with the RNA molecules? They evolved! After just three generations, instead of a single band at the top of the gel, several shorter bands began to appear and ligase activity increased about a thousandfold (see figure). After five generations, ligase activity had further increased and new bands had appeared but became extinct in the ninth generation. In the tenth generation, two major bands had emerged. These were species of RNA that had evolved into ribozymes, and their ligase activity was 7 million times greater than the randomized RNA at the start.
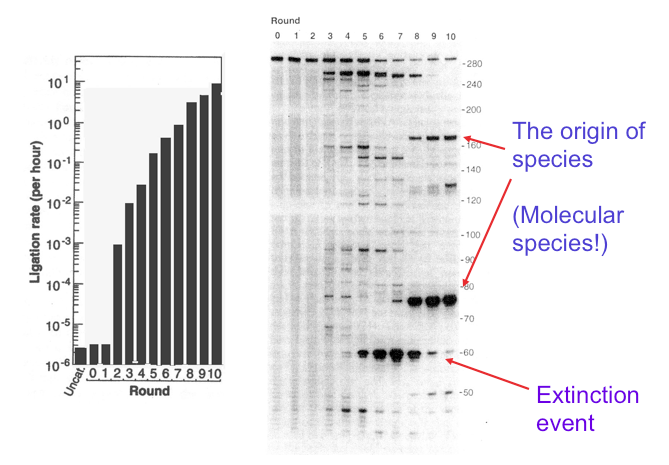
This figure, adapted from Bartel and Szostak’s 1993 paper, shows how genetic information can be introduced into a mixture of random RNA sequences by cycles of selection and amplification. The panel on the left shows catalytic ribozyme ligase activity on a logarithmic scale, and the panel on the right shows how molecular species appear and disappear in the mixture. The lane marked 0 shows the band produced by the original RNA 300 nucleotides in length. At the end of ten rounds, the activity had increased by a factor of 7 million and two bands can be seen, one 75 nucleotides long, and a second about 170 nucleotides in length. These represent the origin of species, but species of molecules, not organisms. Extinction events of other molecular species can also be seen in earlier rounds.
Does this experiment tell us anything about the beginning of evolution? Even though the prebiotic environment was a chaotic mix of chemical reactions driven by various sources of energy, if polymers resembling RNA were synthesized and exposed to cycles of selection and amplification, their base sequences could escape from randomness and incorporate a primitive form of genetic information to become functional catalysts, or ribozymes. But for life to begin, some of those ribozymes had to evolve the ability to catalyze their own reproduction. Does that sound impossible? Eighteen years after Bartel and Szostak published their seminal paper, Phil Holliger reported that his research group had evolved a ribozyme that could act as a polymerase and copy base sequences (Wochner and others 2011).
In just a few more years, I think it is likely that someone will make the convincing claim that they have fabricated a system of molecules that can reproduce itself. This will be the second origin of life, not in nature but in a laboratory. We will never know with certainty how life did begin nearly 4 billion years ago, but we will understand how life can begin.
References
